Copyright 2012 neutronsources.org | All rights reserved. | Powered by FRM II | Imprint / Privacy Policy
Investigating cyan fluorescent proteins for bioimaging with neutron experiments
Date:15.04.19
Source: ill.eu
For the past 25 years, scientists have been developing fluorescent proteins for bioimaging. Today their diversity covers nearly the whole visible spectrum, including blue, cyan, green, yellow, orange, red, far-red and near infrared, allowing scientists to produce detailed images of various proteins’ localization within live cells using multicolour labelling.
A team of researchers at Institut Laue–Langevin (ILL) have used neutron scattering experiments to study a family of fluorescent proteins called Cyan Fluorescent Proteins (CFPs). “CFPs are very useful tools for bioimaging as they can be used to produce pictures of live cells – if the CFP is used as a fusion protein, it can reveal information about protein location, and if used as part of a biosensor, it can report on the evolution of various cell parameters (pH, calcium concentration…)” says Dr Judith Peters, scientist at ILL and professor at Université Grenoble Alpes.
CFPs are variants of Green Fluorescent Protein from the jellyfish Aequorea victoria in which one of the central amino acids in the chromophore – the group responsible for the colour of the compound – has been replaced by a different amino acid, changing the colour from green to cyan. Being able to use multiple colours of fluorescent proteins within a single image is very useful, but it is difficult to achieve optimal fluorescence in cyan fluorescent proteins because the change in amino acid alters hydrogen bonding within the molecule.
This is one of the reasons that scientists have been working hard to improve and enhance CFPs for many years. It has been a challenging task and at one point there was a 13-year gap between the availability of the first useable CFP and the discovery of a variant with almost perfect fluorescence efficiency.
In order to be useful as biomarkers, CFPs must emit sufficient fluorescent light to be easily and reliably detectable. The fluorescence efficiency (or quantum yield) must be as high as possible and it can be affected by the molecular dynamics of proteins, such as the flexibility or rigidity of the protein core.
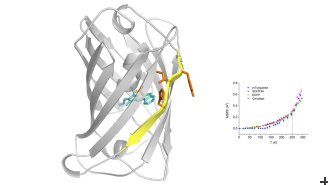
The researchers used a technique at ILL called incoherent neutron scattering to look at the molecular dynamics of five different CFPs. By firing beams of neutrons at the CFPs and carefully analysing the pattern of scattered neutrons, scientists were able to observe how atoms and molecules within the proteins vibrate, rotate and move, revealing crucial information about their dynamic properties. Neutron experiments are a good method for investigating the dynamics of proteins because they are non-invasive and do not damage the proteins. Other techniques often involve higher energy which can destroy the samples.
“Our works builds on previous experiments done by Antoine Royant and his team at Institut de Biologie Structurale, which investigated which parts of the proteins are moving,” says Dr Peters. This work revealed that the protein motions were mainly connected to a part of the protein core which is organized as a ß-barrel – specifically to the seventh strand of this barrel.
The ILL team’s experiments showed that there was an increase in flexibility of the protein core between the first usable CFP, called enhanced cyan fluorescent protein (ECFP), and the second variant, called Cerulean. This flexibility can actually positively affect fluorescence efficiency. However, in the next three variants that scientists developed, called SCFP3A, mTurquoise and mTurquoise2, this flexibility was eliminated, and these proteins showed greater rigidity and additional fluorescence efficiency compared to Cerulean. This confirms that increasing the rigidity of the direct environment of the fluorescent chromophore is not the sole parameter leading to brighter fluorescent proteins and that increased flexibility in some cases may be helpful. The best CFP variant for bio-imaging is mTurquoise2, which has the most rigid protein core of all.
Collecting data on protein dynamics is thus useful for scientists who are working to develop improved fluorescent proteins in a range of colours for bio-imaging. “The next step is to repeat these experiments under high pressure and to observe how this affects protein dynamics,” says Dr Peters.
Re.: “Journal of the Royal Society interface, March 2019”:“https://royalsocietypublishing.org/doi/10.1098/rsif.2018.0848. “ Doi:10.5291/ILLDATA.8-04-727”:https://royalsocietypublishing.org/doi/10.1098/rsif.2018.0848
See also doi:10.5291/ILL-DATA.8-04-761.
ILL instruments: Elastic incoherent neutron scattering (EINS) experiments were led on spectrometers IN6, IN13 and IN16B.