Copyright 2012 neutronsources.org | All rights reserved. | Powered by FRM II | Imprint / Privacy Policy
Neutrons paint atomic portrait of prototypical cell signaling enzyme—protein kinase A
Date: 21.03.2019
Source: ill.eu
Direct observations of the structure and catalytic mechanism of a prototypical kinase enzyme—protein kinase A or PKA—will provide researchers and drug developers with significantly enhanced abilities to understand and treat fatal diseases and neurological disorders such as cancer, diabetes, and cystic fibrosis.
The discovery was made by an international team of researchers using macromolecular neutron crystallography at the Department of Energy’s Oak Ridge National Laboratory (ORNL) and the Institut Laue-Langevin (ILL) in Grenoble, France. Building on a decade-long investigation, the combined efforts revealed previously unknown characteristics of the archetypal protein kinase, including a complete map of the atomic structure and the underpinnings of the enzyme’s chemistry used in cellular signaling. The details are published in the journal Science Advances.
Kinases are a large group of hundreds of enzymes responsible for initiating and regulating a myriad of cellular processes. Kinases send signals to proteins by way of phosphorylation—essentially adding a reactive chemical group consisting of phosphorus and oxygen to a specific location on a substrate protein to activate its specific biological function.
Problems arise when gene mutations occur that cause kinases to work improperly, which in turn leads to disease. For example, a malfunctioning kinase, or one that can’t be deactivated, could be responsible for the overexpression of proteins that lead to the uncontrollable propagation of cancer cells. The opposite is also true in neurological disorders in which a malfunctioning kinase simply causes cells to die.
“Creating drugs that target specific kinases is extremely important,” said ORNL co-author Andrey Kovalevsky. “This detailed understanding of the PKA structure and its dynamics will tell us more about other kinds of kinases and should help drug developers design new drugs with better specificity, which would ultimately translate to more precise therapeutics with fewer side effects.”
Changes in the protein structure due to mutations can be difficult to detect using x-ray scattering, even though the effects of mutations on the protein dynamical behavior can be quite profound. However, these dynamical behaviors become more apparent by examining the overall hydrogen bonding network throughout the protein.
Neutrons are uniquely sensitive to light elements such as hydrogen. Because approximately 50% of all atoms in proteins are hydrogen, and most of the chemical reactions that enzymes catalyse involve hydrogen, neutrons are an ideal and unsurpassed probe for detecting the positions of hydrogen atoms within the protein structure and tracking their movement during catalysis.
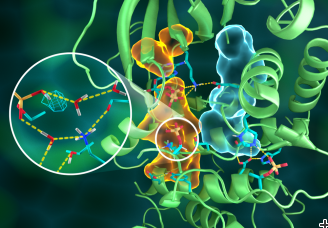
Preliminary neutron diffraction tests were made using the IMAGINE diffractometer at ORNL’s High Flux Isotope Reactor with full data collection performed using the LADI-III diffractometer at ILL. These data enabled researchers to see the exact positions of hydrogen atoms to reveal the complete kinase structure as well as the protonation states of all amino acids in PKA and the ligands bound to its active site, many of which were previously unknown.
Matthew Blakeley, LADI-III beamline scientist at the ILL and co-author of the study, said “Given the relatively large unit-cell of PKA and the fairly small crystal volumes available (0.1mm3), neutron diffraction data collection to sufficiently high-resolution to reveal the protonation states of PKA was only possible using the LADI-III beamline at ILL, and therefore emphasises the importance of international scientific collaborations.”
“Neutrons enabled us to validate x-ray predictions of hydrogen atom positions, as well as determining the protonation states of chemical groups that were not known, or in places where we didn’t expect to find them. This major chemical advance paves the way for more sophisticated molecular modeling and simulation studies of protein kinase structure and function.” said University of California–San Diego corresponding author Susan Taylor.
“Neutrons offer an unprecedented view of the hydrogen bond network surrounding and within the active site of PKA and give crucial insights on how allosteric transmission of information through the protein structure occurs,” added University of Minnesota co-author Gianluigi Veglia. “Combining neutron crystallography with nuclear magnetic resonance spectroscopy will provide a comprehensive understanding of how enzymes work.”
Re. Gerlits et al., Sci. Adv., 2019; 5: eaav0482. DOI: 10.1126/sciadv.aav0482
Contact: Dr M. Blakeley, ILL